Kiedrowki’s group has constructed a chemical self-replicating system (SBK) with autocatalytic parabolic growth which can be observed via fluorescence [1]. It was used as model chemistry for the amplification of information contents in artificial cells in microfluidic structures, prior to the availability of coupled lipid-template systems. The replication can be monitored in real-time by fluorescence resonance transfer (FRET). In addition, and for comparison, an enzymatic isothermal amplification system was also employed: Strand Displacement Amplification (SDA). This reaction, originally proposed by Walker [2], has been studied extensively in our laboratory. It utilizes a DNA polymerase and restriction enzyme together with modified nucleotides and short DNA primers, to nick one strand of a DNA duplex and polymerize fresh DNA while displacing the nicked strand, thereby avoiding the double stranded endpoint of the simple DNA polymerization process. Details may be found in [3, 4].
Firstly, the SBK system was reconstituted and tested on a "normal" lab scale, i.e. 50 µl sample volume. The reaction kinetics were monitored by FRET (Cy5-Cy3) in a fluorescence spectrophotometer. The product formation can be seen by a decrease in fluorescence intensity, with the reaction initiated by the addition of buffered EDC. All experiments use the pentameric oligonucleotide system: the 3’-phosphate as well as the 5’-amino building blocks were five nucleotides long and the template assembled has a length of ten nucleotides. For further details see Schöneborn [5].
The influence of different initial template concentrations as starting material (1-10 mol % of the concentration of the building blocks) was tested and in addition the reaction without "external template" monitored. After the reaction chemistry was established at BioMIP, the system could be implemented in the "fan"-reactor.
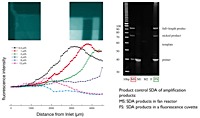
Performing SDA in the fan reactor leads to successful formation of specific amplification products (see also upper left figure). Varying the overall inflow rate to the fan reactor shifts this region but stability of a stationary replication standing wave front is maintained. This "homeostatic" standing wave provides a suitable object of study for long-term replication and evolution experimentation in connection with self-replication en route to artificial cells. We also took advantage of the symmetry of the "fan"-reactor to solve reaction-diffusion equations for the relaxation and spatial dependence of the non-equilibrium standing wave, and compared these results with experimental profiles captured via fluorescence video microscopy for both chemical test reactions. The results establish that, in the microfluidic fan reactor, chemical amplification reactions can be sustained robustly under constant conditions using comparatively small volumes of reactants. The opposite 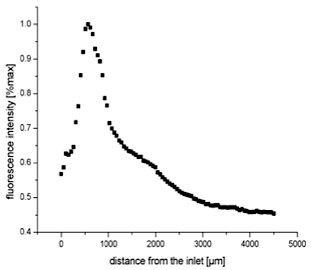 figure shows the ongoing spatially resolved on-line detection of chemical self-replication system (SBK) in the "fan"-reactor.
figure shows the ongoing spatially resolved on-line detection of chemical self-replication system (SBK) in the "fan"-reactor.
The plot shows the decrease in fluorescence
intensity associated with the creation of replication products, as a function of radial distance
from the inlet to the fan reactor.