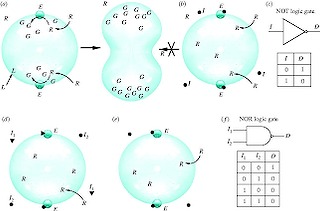
As part of CIRAS involvement in WP2, we started an ambitious exploration of potential scenarios for protocell computation as an integrative part of our understanding of protocell dynamics. Our interest started with the potential use of self-reproduction as part of the computational process (see figure) and how non-standard forms of computation would emerge. During our study, we also addressed the problem of the potential limits of protocell-based computing resulting from spatial and dynamical constrains. In this context, we were interested in looking at the problems arising from the implementation of arbitrarily complex logic devices. Specifically, we were interested in how (if possible) to overcome the limitations imposed by the standard procedure of mapping cellular and digital circuits. During this process we found that there is a totally different, and systematic form of approaching the problem that largely avoids the problem of the wiring by distributing computations and cell-cell communication in the appropriate way. A general algorithm has been designed (able to find and simplify the desired circuits) and several nontrivial case studies have been shown to be implementable in engineered cells (specific designs for E. coli have been obtained). This is currently being a major area of research and will benefit from the join efforts made bith within PACE and our new EU project CELLCOMPUT.
RELATED PUBLICATIONS
Ricard V. Solé, Andreea Munteanu, Carlos Rodriguez-Caso and Javier Macía, "Synthetic protocell biology: from reproduction to computation", Phil. Trans. Roy. Soc. Lond. ser. B 362, 1727-1739 (2007).