 At an early stage in pre-biotic evolution, groups of replicating
molecules must have coordinated their reproduction to form aggregated units of
selection. Mechanisms that enable this to occur are currently not well
understood. This raises the question of how, and when, the organism as a whole
can be viewed as a unit of selection. This is a necessary condition for such
systems to evolve into
contemporary organisms, with a well-defined separation between the genotype and
phenotype, and a coordinated replication. To gain understanding of these
phenomenon we have studied a deterministic model of primitive replicating
aggregates, proto-organisms, that host populations of replicating information
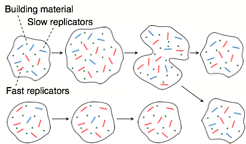
carrying molecules. Some of the molecules promote the reproduction of the
proto-organism (blue strains in the picture to the right) at the cost of their
individual replication rate compared to faster replicators (red in the
picture). A situation resembling that of group selection arises. We have derive and analytically solve
a partial differential equation
that describes the system. The main finding is that the relative prevalence of
fast and slow replicators is determined by the relative strength of selection
at the aggregate level to the selection strength at the molecular level. We
have also made some analysis of finite population size effects.
At an early stage in pre-biotic evolution, groups of replicating
molecules must have coordinated their reproduction to form aggregated units of
selection. Mechanisms that enable this to occur are currently not well
understood. This raises the question of how, and when, the organism as a whole
can be viewed as a unit of selection. This is a necessary condition for such
systems to evolve into
contemporary organisms, with a well-defined separation between the genotype and
phenotype, and a coordinated replication. To gain understanding of these
phenomenon we have studied a deterministic model of primitive replicating
aggregates, proto-organisms, that host populations of replicating information
carrying molecules. Some of the molecules promote the reproduction of the
proto-organism (blue strains in the picture to the right) at the cost of their
individual replication rate compared to faster replicators (red in the
picture). A situation resembling that of group selection arises. We have derive and analytically solve
a partial differential equation
that describes the system. The main finding is that the relative prevalence of
fast and slow replicators is determined by the relative strength of selection
at the aggregate level to the selection strength at the molecular level. We
have also made some analysis of finite population size effects.
In more detail our model focus on the evolutionary dynamics of systems consisting of self-assembling container aggregates that contain populations of self-replicating information carrying molecules -proto-genes. The aggregates should be viewed as primitive proto-organisms, with genomes consisting of evolving populations of proto-genes. The aggregates grow by continuously incorporating new building blocks. At a certain size they become unstable and spontaneously divide, at which point, we say that the proto-organism has replicated. To mimic the chemical setup for the minimal proto-cell in the PACE project, the production of new building blocks is catalyzed by the proto-genes, for example through an electron charge transfer process. A genome’s ability to self-replicate and its catalytic affect on the growth of the aggregate are assumed to be uncorrelated. Since the sequence space in very large we can then assume that a sequence is very unlikely to be optimal for both auto-catalysis and aggregate growth. The result is that certain strains of proto-genes are efficient as self-replicators, whereas other strains are more active in the catalysis of new building blocks and contribute more to the reproduction of the container. The evolution of the system as a whole is then characterized by a conflict reminiscent of group selection.
Technically we we use the following logistic growth model for the internal population dynamics, where x denotes the relative concentration of fast replicators in the aggregate:
![]()
![]()
The growth of the aggregate is proportional to how many slow
replicators there are in the aggregate:
![]()
![]()
The evolution the probability distribution of slow and fast replicators can now be described by the following PDE:
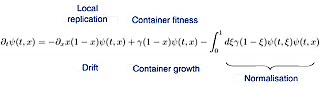
 where the important dimensionless
factor is defied as
where the important dimensionless
factor is defied as
![]()
![]()
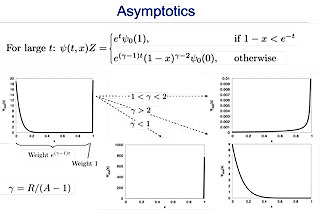
The model can be solved analytically but the important conclusions come from studying the asymptotic behavior of the model. The asymptotic behavior of the model:
The main conclusion is that there are two main regions in which the model shows different behavior:
R<2A : Fast replicators completely dominate the population.
R>2A : Slow replicators dominate the population.
Reference: